Explore the complete implementation: GitHub Repository
Problem Statement
This project addresses the challenge of classifying brain tumor types from MRI images using pretrained CNN architectures. Our goal is to benchmark performance across popular transfer learning models under standardized conditions, focusing on diagnostic accuracy and generalization across glioma, meningioma, and pituitary tumors.
Model Performance Summary
| Model | Test Accuracy | Precision | Recall | Notes |
|---|---|---|---|---|
| Xception | 88.18% | 90.10% | 86.04% | Best performance overall |
| MobileNetV2 | 84.59% | 86.05% | 83.30% | Some overfitting |
| EfficientNetB0 | 30.89% | 0.00% | 0.00% | Complete training failure |
Why This Matters
Before diving into the technical implementation of this project, it's essential to understand why this work matters. Brain tumor detection is not just a computational challenge, it’s a life critical task with real world implications. The types of tumors discussed here differ dramatically in prognosis, treatment approach, and urgency. By establishing a clear clinical picture of these conditions, we can better appreciate the role that machine learning can play in supporting early, accurate, and scalable diagnosis.
Clinical Challenge: The Need for Automated Diagnostics
Brain tumors represent one of the most critical diagnostic challenges in modern medicine. These abnormal cell proliferations can be benign or malignant, with early detection being paramount for successful treatment outcomes and patient survival rates.
While Magnetic Resonance Imaging (MRI) provides exceptional soft tissue contrast for tumor visualisation, the manual interpretation process faces significant bottlenecks:
- Time Constraints: Radiologists may review 100+ scans daily, leading to fatigue-induced oversights
- Diagnostic Variability: Inter-observer agreement rates can vary significantly between practitioners
- Resource Limitations: Many healthcare systems face critical shortages of specialised radiologists
- Urgent Triage Needs: Emergency cases require rapid preliminary assessments
This project investigates how Convolutional Neural Networks (CNNs) can serve as intelligent diagnostic support tools, providing rapid preliminary assessments while maintaining the essential human expertise in final diagnosis.
According to the World Health Organisation (WHO), a full clinical diagnosis of brain tumors includes identifying not only the tumor’s type but also its grade and malignancy level.
In this project, we focus only on classifying the type of tumor based on MRI features, not on grading or malignancy.
This is a crucial step in building automated systems that could eventually aid in faster and more scalable triage, particularly in resource-limited settings.
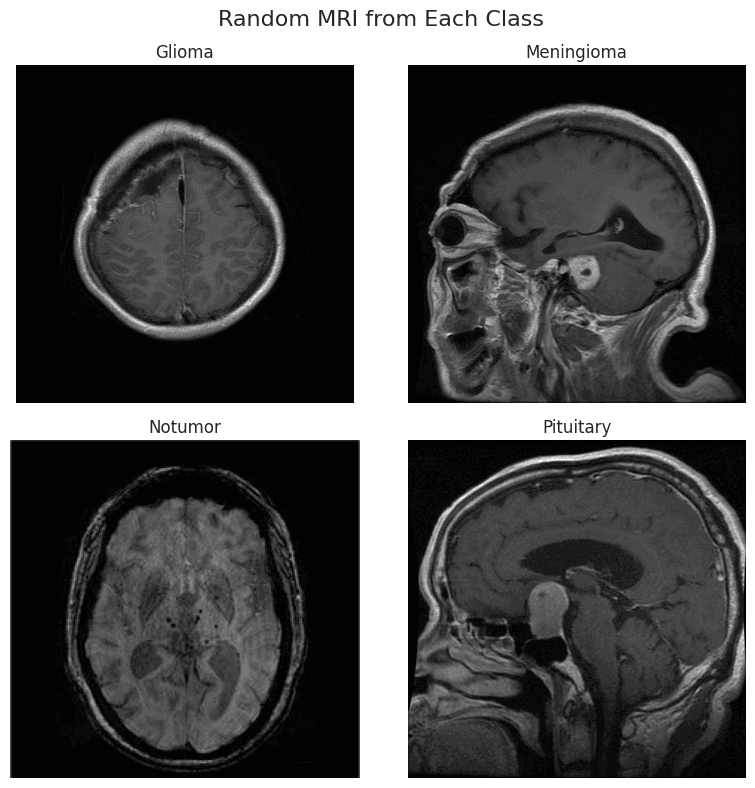
Tumor Types in This Study
- Pituitary Adenomas (Often benign & highly treatable)
Location: Sellar and suprasellar regions, the “master gland” area behind the eyes.
Characteristics: Benign tumors from the pituitary gland, often hormonally active.
Clinical Impact: Can lead to Cushing’s disease, vision loss, mood swings, and fertility problems due to hormonal disruption and optic chiasm compression.
MRI Features: Well-defined masses with strong post-contrast enhancement.
- Gliomas (Median survival < 15 months)
Location: Can appear in cerebral hemispheres, brainstem, or cerebellum.
Characteristics: Range from low-grade to aggressive glioblastomas (Grade IV).
Clinical Impact: Symptoms depend on location — seizures, personality change, speech difficulties, or weakness. Glioblastomas have a median survival under 15 months.
MRI Features: Irregular infiltrative growth with peritumoral edema and mass effect.
- Meningiomas (Slow-growing, usually noncancerous)
Location: Along dural surfaces from arachnoid cap cells (outer brain lining).
Characteristics: Most common adult brain tumor, usually benign and slow-growing.
Clinical Impact: Symptoms arise from compression, headaches, cognitive changes, seizures depending on tumor position.
MRI Features: Homogeneous enhancement with the classic “dural tail” sign.
Why Use CNNs?
Convolutional Neural Networks (CNNs) are designed to analyse visual data. They work by applying filters that detect patterns in an image, starting from simple edges and shapes, then building toward complex features like tumour textures and outlines.
This layered pattern recognition makes CNNs particularly effective for medical image classification, such as identifying brain tumours from MRI scans.
What Are Pretrained Models?
Pretrained models are neural networks that have been previously trained on large and diverse datasets, such as ImageNet, which contains over a million natural images across a thousand categories.
These models have already learned how to extract rich and generalisable visual features like edges, textures, and object parts.
In this project, using pretrained backbones like Xception, MobileNetV2, and EfficientNetB0 allows the model to leverage these learned features as a starting point, dramatically reducing training time and improving convergence.
Instead of learning from scratch, we fine-tune these models on our specific dataset of brain MRI images. This approach is especially useful when working with limited labelled medical data, where training a large model from the ground up would be impractical or prone to overfitting.
Display of MRI Images
Dataset & Scope of Classification
This project uses a publicly available brain MRI dataset that consolidates images from three sources: Figshare, SARTAJ, and Br35H. It contains 7,023 brain MRI images across four categories: glioma, meningioma, pituitary, and no tumor.
Note: The glioma samples from the SARTAJ dataset were found to be mislabeled and were replaced with verified images from Figshare to ensure data integrity.
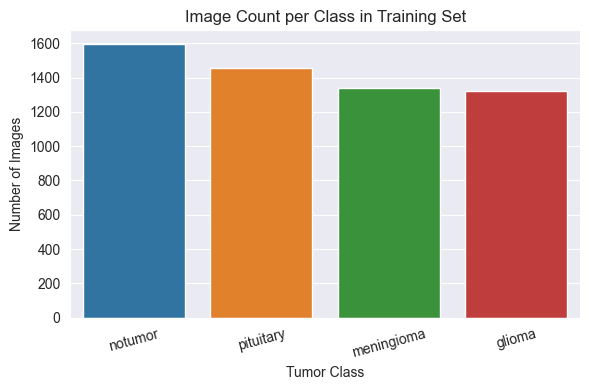
Training Set Distribution
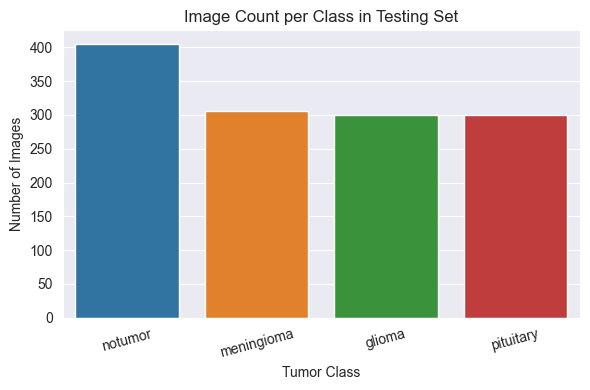
Test Set Distribution
Although the dataset is generally balanced across the four tumour types, the no tumor class is slightly overrepresented in both the training and testing sets.
This minor imbalance was not sufficient to warrant the use of class weighting or sampling adjustments, but it may influence evaluation metrics such as recall or precision.
Preprocessing
The training set was divided using stratified sampling to ensure balanced class representation across training and validation subsets. All images were resized to 224×224 pixels for input compatibility with pretrained CNNs.
To enhance generalisation and reduce overfitting, data augmentation was applied to the training set using TensorFlow’s ImageDataGenerator, including random rotations (±10°), zoom transformations (±10%), and horizontal flips. Since the dataset included multiple perspectives (axial, sagittal, and coronal), horizontal flipping was deemed appropriate for regularisation.
Validation and test sets were not augmented and only rescaled to the [0, 1] range, preserving evaluation integrity. This preprocessing approach ensured consistency and robustness across all models during training and evaluation.
Development Environment & Model Approach
Compute Environment: Trained in Google Colab with Tesla T4 GPU acceleration
Model Type: Deep learning classifier using Convolutional Neural Networks (CNNs)
Pretrained Backbones: MobileNetV2, EfficientNetB0, and Xception, all initialised with ImageNet weights
Input Dimensions: All MRI slices resized to 224×224 pixels
Frameworks Used: TensorFlow 2.x with the Keras high-level API
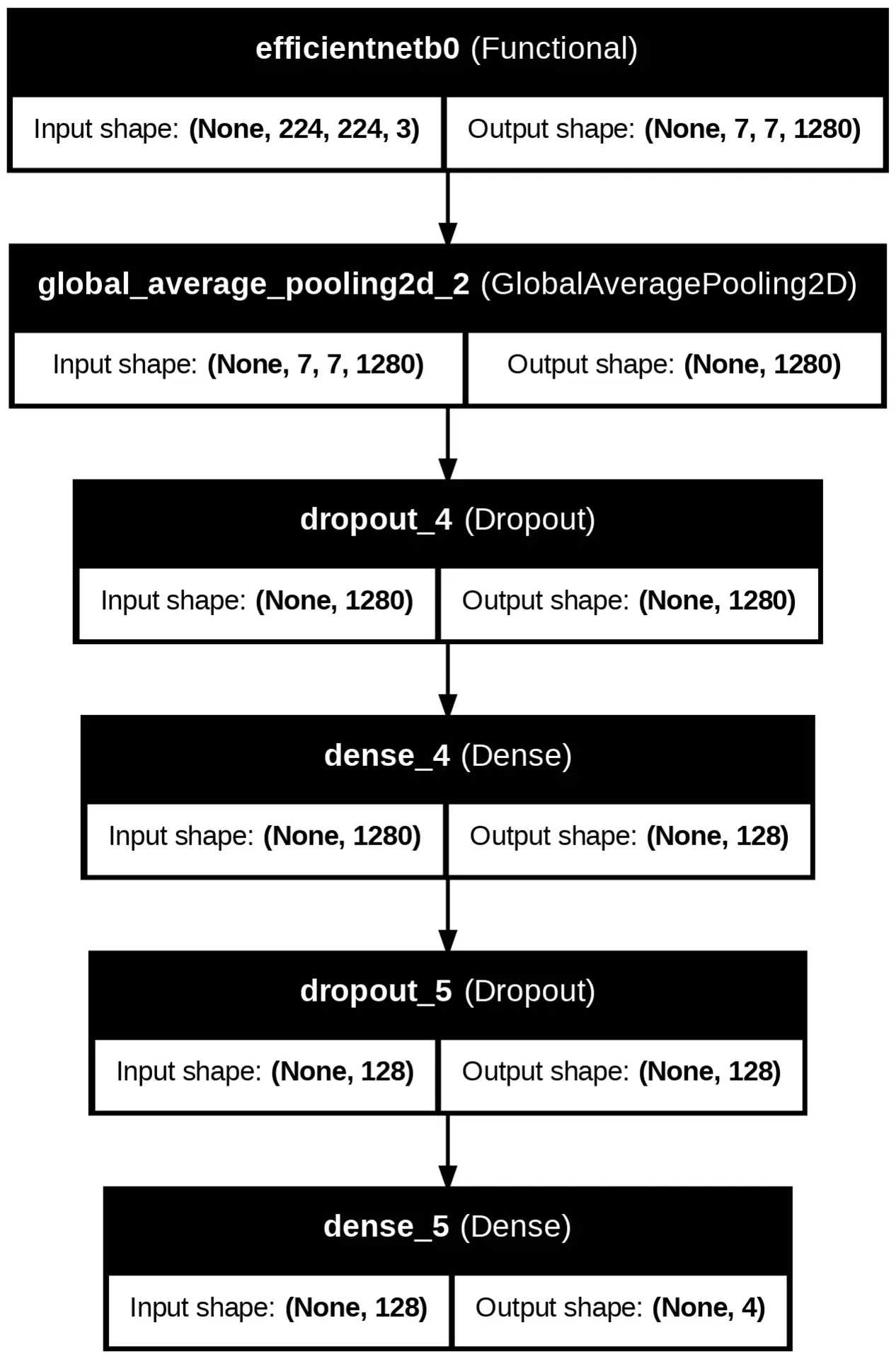
All three models, MobileNetV2, EfficientNetB0, and Xception, share the same classification head and training configuration. The only difference between them lies in the pretrained backbone used for feature extraction.
All input images were resized to 224×224 pixels for consistency across models.
However, internal feature map sizes differed based on the architecture.
For example, Xception produced a deeper output feature map of shape (7, 7, 2048) compared to the other models.
The following animation visualises the internal structure of each model architecture used in this experiment.
Model Performance
Three pretrained CNN models were evaluated on the brain MRI classification task: MobileNetV2, EfficientNetB0, and Xception. Each model was trained for 10 epochs on the same dataset under identical conditions, allowing for a direct comparison of their learning capabilities and generalisation performance.
Model Performance Summary
| Model | Train Accuracy | Val Accuracy | Test Accuracy | Test Precision | Test Recall |
|---|---|---|---|---|---|
| Xception | 91.81% | 90.11% | 88.18% | 90.10% | 86.04% |
| MobileNetV2 | 92.67% | 87.66% | 84.59% | 86.05% | 83.30% |
| EfficientNetB0 | 27.93% | 27.91% | 30.89% | 0.00% | 0.00% |
Visual Results
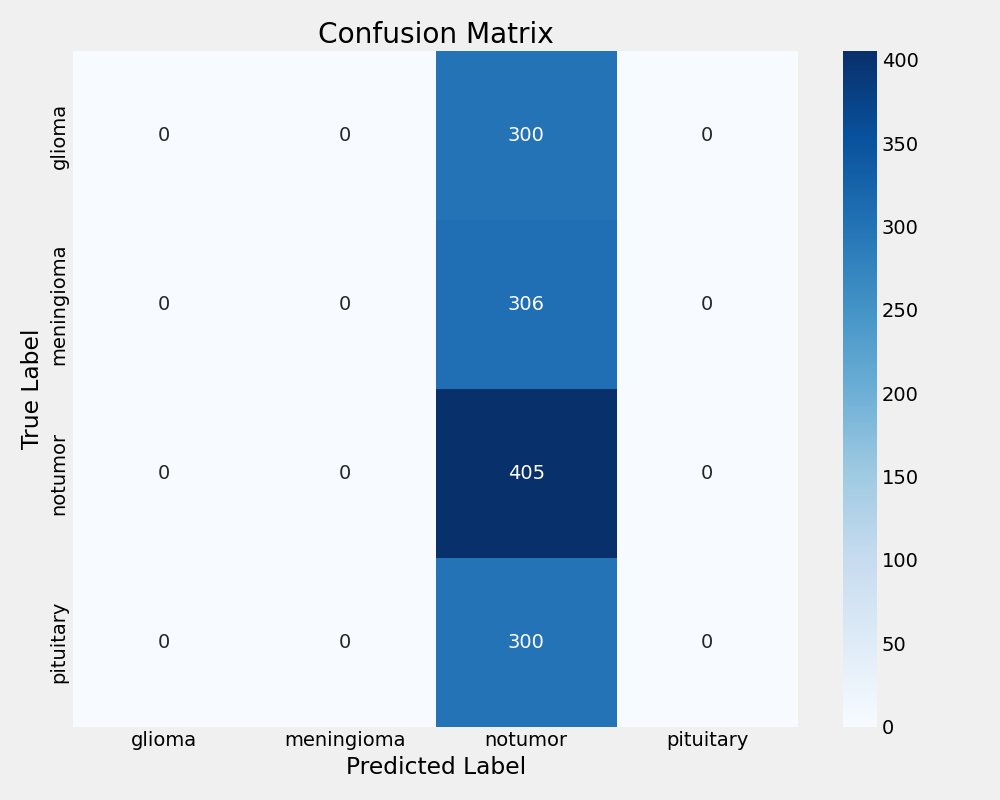
EfficientNetB0 Confusion Matrix
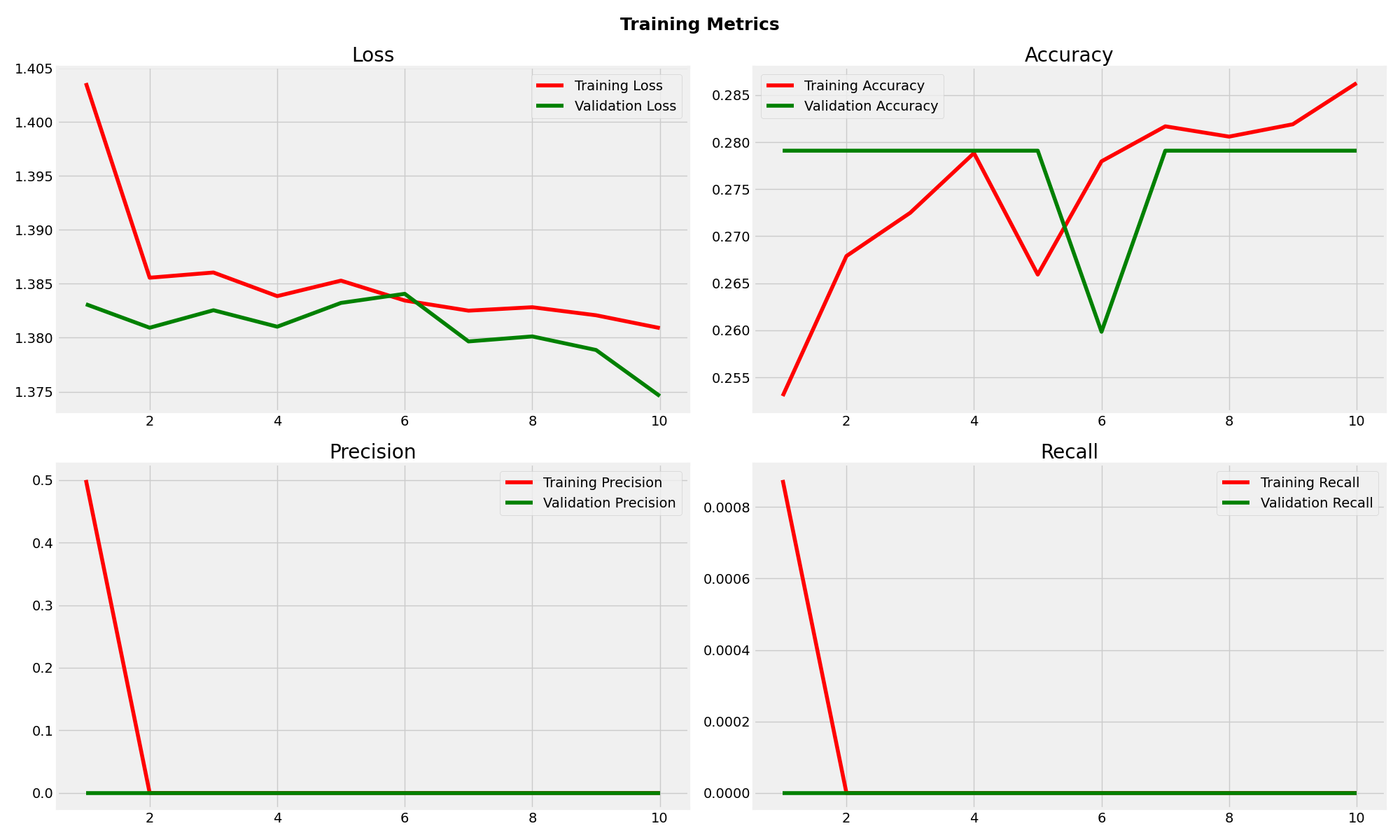
EfficientNetB0 Training Metrics
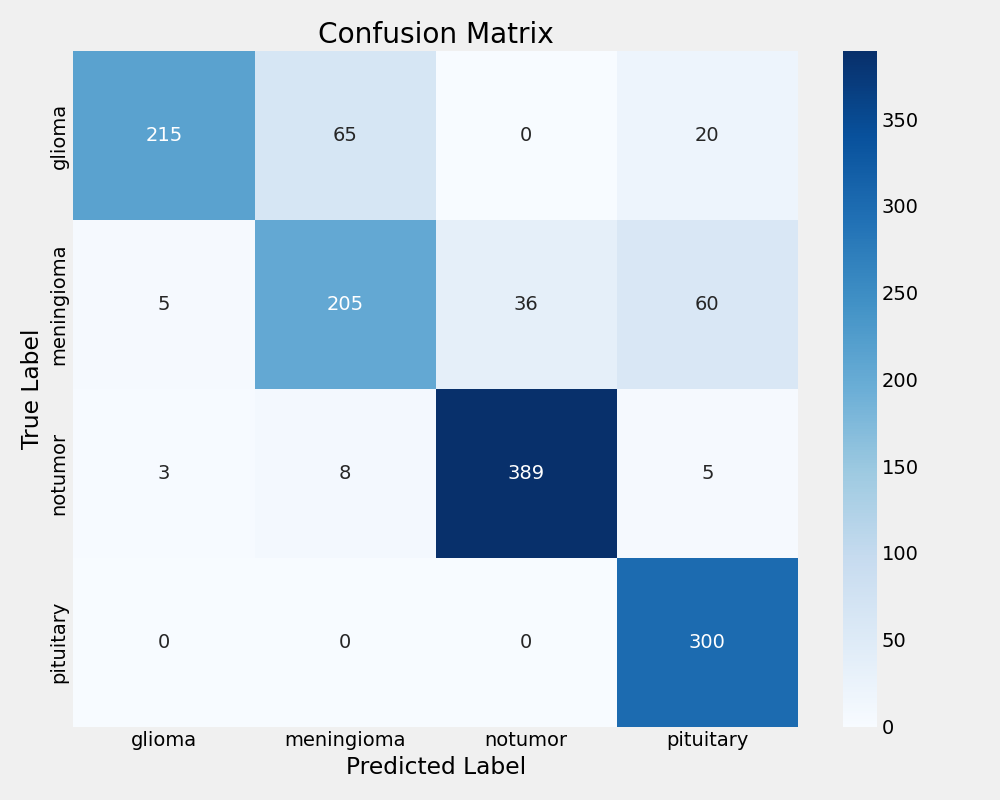
MobileNetV2 Confusion Matrix
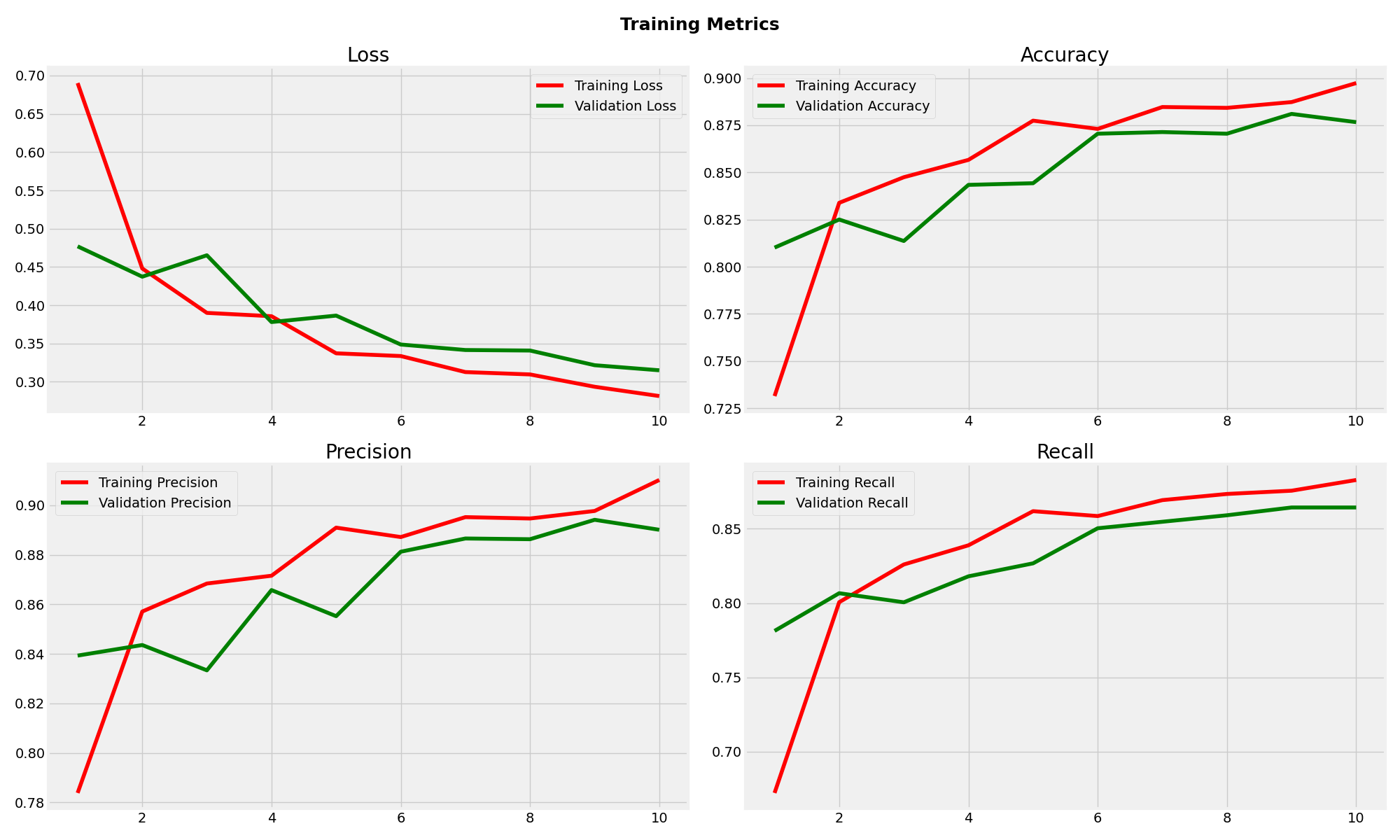
MobileNetV2 Training Metrics
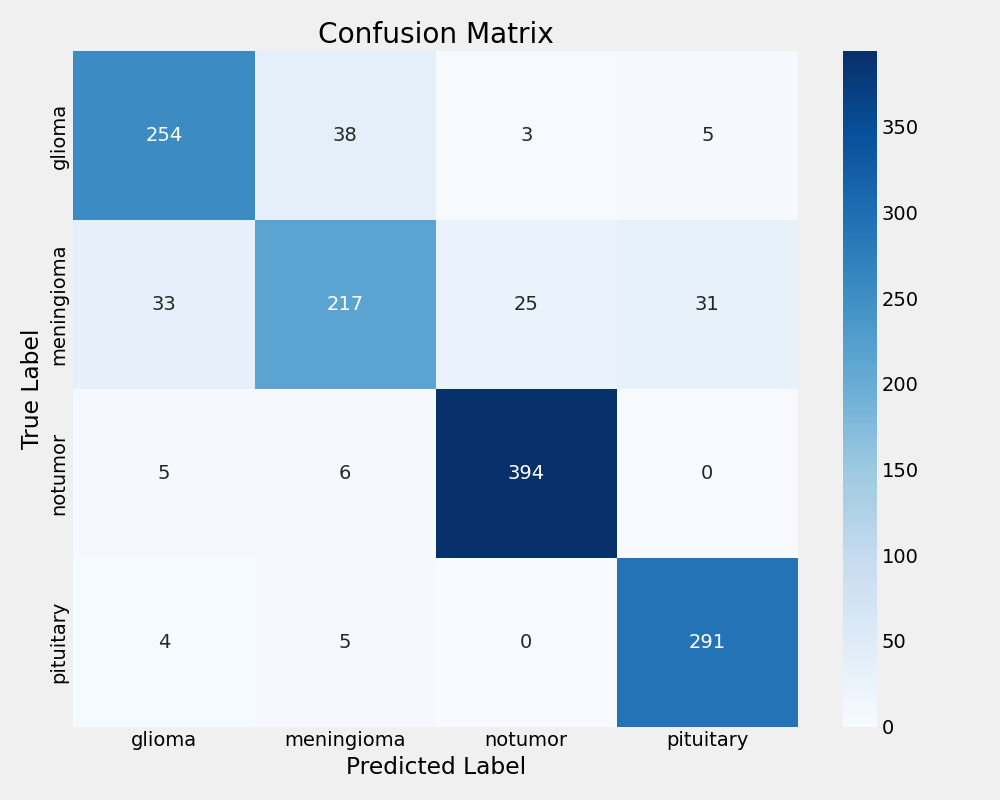
Xception Confusion Matrix
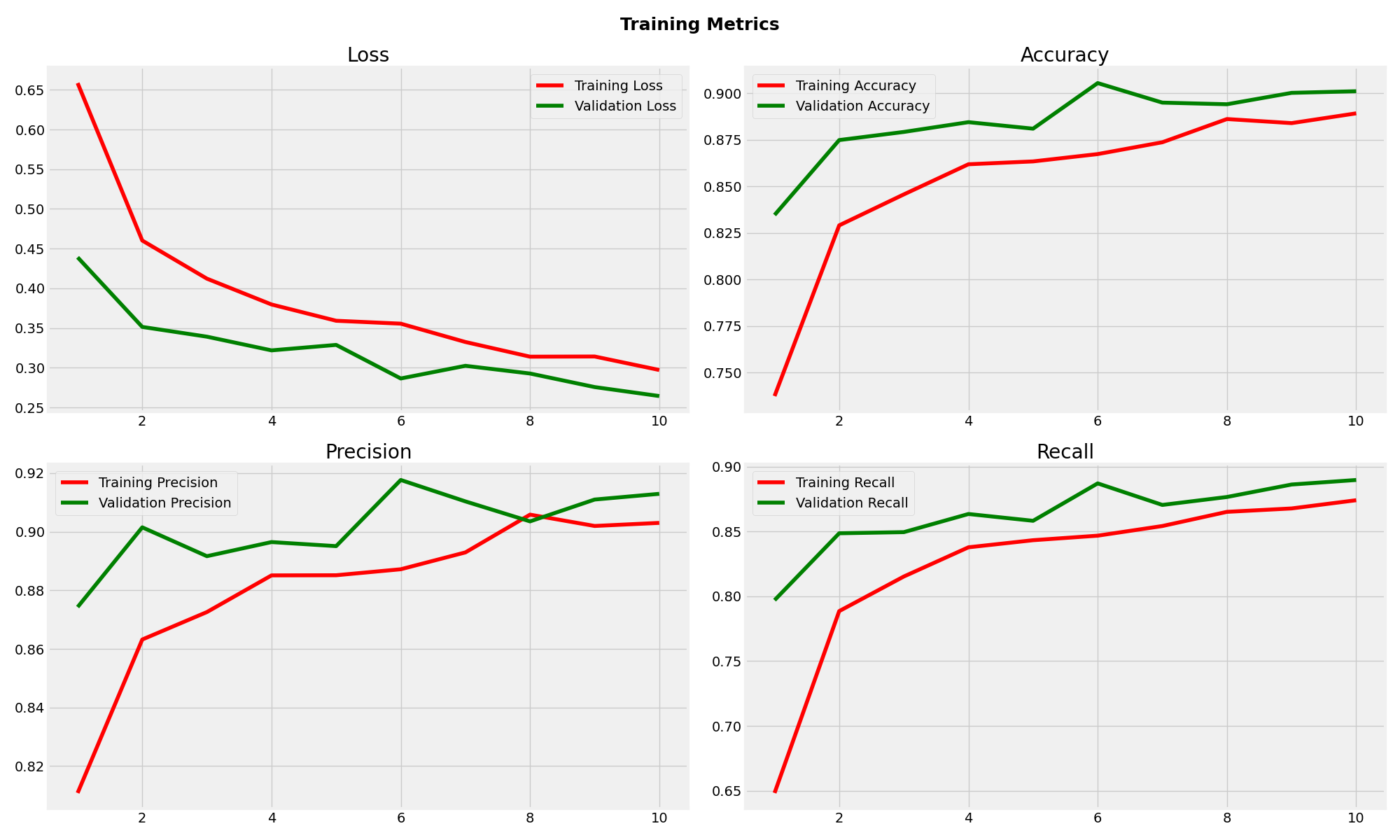
Xception Training Metrics
Observations
Xception demonstrated the most robust performance with 88.18% test accuracy and excellent generalisation. The minimal gap between training (91.81%) and test accuracy indicates strong model stability. With balanced precision (90.10%) and recall (86.04%), Xception showed consistent classification across all tumor types, making it the most suitable choice for clinical deployment.
MobileNetV2 achieved reasonable performance with 84.59% test accuracy but exhibited concerning overfitting patterns. The significant gap between training (92.67%) and validation (87.66%) accuracy suggests limited generalisation capability. While precision remained acceptable at 86.05%, the lower recall (83.30%) indicates potential missed classifications in clinical scenarios.
EfficientNetB0 experienced complete training failure with 30.89% test accuracy, performing worse than random classification. The model achieved zero precision and recall, consistently predicting only the "notumor" class regardless of input. This failure highlights the critical importance of base model selection in transfer learning applications and demonstrates that architectural efficiency does not guarantee task suitability.
The comparative analysis reveals that model architecture significantly impacts transfer learning effectiveness for medical imaging tasks. Xception's superior performance across all metrics establishes it as the optimal choice, while EfficientNetB0's failure provides valuable insights into the limitations of assuming universal model applicability.
Consideration
While Xception demonstrated superior performance in this experiment, it is important to note that with additional hyperparameter tuning, architecture modifications, and training optimisation, both MobileNetV2 and EfficientNetB0 could potentially achieve comparable results.
Each pretrained backbone offers unique strengths, MobileNetV2 for its computational efficiency, EfficientNetB0 for its compound scaling strategy, and Xception for its use of depthwise separable convolutions. These characteristics can be further leveraged through targeted optimisation techniques tailored to each model.
